SMOOTHN - Robust spline smoothing for 1-D to N-D data
SMOOTHN provides a fast, automatized and robust discretized spline smoothing for data of arbitrary dimension.
Z = SMOOTHN(Y) automatically smoothes the uniformly-sampled array Y. Y can be any N-D noisy array (time series, images, 3D data,...). Non finite data (NaN or Inf) are treated as missing values.
Z = SMOOTHN(Y,S) smoothes the array Y using the smoothing parameter S. S must be a real positive scalar. The larger S is, the smoother the output will be. If the smoothing parameter S is omitted (see previous option) or empty (i.e. S = []), it is automatically determined by minimizing the generalized cross-validation (GCV) score.
Z = SMOOTHN(Y,W) or Z = SMOOTHN(Y,W,S) smoothes Y using a weighting array W of positive values, which must have the same size as Y. Note that a nil weight corresponds to a missing value.
[Z,S] = SMOOTHN(...) also returns the calculated value for the smoothness parameter S so that you can fine-tune the smoothing subsequently if needed.
Contents
- Smoothing vector fields or multi-component data
- Robust smoothing
- Different spacing increments
- Example #1: smooth a curve
- Example #2: smooth a surface
- Example #3: smooth 2-D data with missing values
- Example #4: smooth volumetric data
- Example #5: smooth a cardioid
- Example #6: smooth a 3-D parametric curve
- Example #7: smooth a 2-D vector field
- Example #8: smooth a 3-D vector field
- Pick of the week in Matlab FEX
- References
- See also
- About the author
Smoothing vector fields or multi-component data
If you want to smooth a vector field or multicomponent data, Y must be a cell array. For example, if you need to smooth a 3-D vectorial flow (Vx,Vy,Vz), use Y = {Vx,Vy,Vz}. The output Z is also a cell array which contains the smoothed components. See examples below.
Robust smoothing
Z = SMOOTHN(...,'robust') carries out a robust smoothing that minimizes the influence of outlying data.
An iteration process is used with the 'ROBUST' option, or in the presence of weighted and/or missing values. Z = SMOOTHN(...,OPTIONS) smoothes with the termination parameters specified in the structure OPTIONS. OPTIONS is a structure of optional parameters that change the default smoothing properties. It must be the last input argument.
The structure OPTIONS can contain the following fields:
- OPTIONS.TolZ: Termination tolerance on Z (default = 1e-3), OPTIONS.TolZ must be in ]0,1[
- OPTIONS.MaxIter: Maximum number of iterations allowed (default = 100)
- OPTIONS.Initial: Initial value for the iterative process (default = original data, Y)
- OPTIONS.Weight: Weight function for robust smoothing: 'bisquare' (default), 'talworth' or 'cauchy'
[Z,S,EXITFLAG] = SMOOTHN(...) returns a boolean value EXITFLAG that describes the exit condition of SMOOTHN; 1: SMOOTHN converged, 0: Maximum number of iterations was reached.
Different spacing increments
SMOOTHN, by default, assumes that the spacing increments are constant and equal in all the directions (i.e. dx = dy = dz = ...). This means that the smoothness parameter is also similar for each direction. If the increments differ from one direction to the other, it can be useful to adapt these smoothness parameters. You can thus use the following field in OPTIONS:
- OPTIONS.Spacing = [d1 d2 d3...], where dI represents the spacing between points in the Ith dimension.
Important note: d1 is the spacing increment for the first non-singleton dimension (i.e. the vertical direction for matrices).
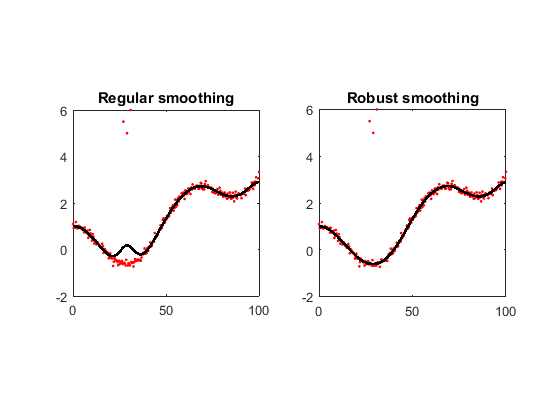
Example #1: smooth a curve
x = linspace(0,100,2^8); y = cos(x/10)+(x/50).^2 + randn(size(x))/10; y([70 75 80]) = [5.5 5 6]; z = smoothn(y); % Regular smoothing zr = smoothn(y,'robust'); % Robust smoothing subplot(121), plot(x,y,'r.',x,z,'k','LineWidth',2) axis square, title('Regular smoothing') subplot(122), plot(x,y,'r.',x,zr,'k','LineWidth',2) axis square, title('Robust smoothing')
Example #2: smooth a surface
xp = 0:.02:1; [x,y] = meshgrid(xp); f = exp(x+y) + sin((x-2*y)*3); fn = f + randn(size(f))*0.5; fs = smoothn(fn); subplot(121), surf(xp,xp,fn), zlim([0 8]), axis square subplot(122), surf(xp,xp,fs), zlim([0 8]), axis square

Example #3: smooth 2-D data with missing values
n = 256; y0 = peaks(n); y = y0 + randn(size(y0))*2; I = randperm(n^2); y(I(1:n^2*0.5)) = NaN; % lose 1/2 of data y(40:90,140:190) = NaN; % create a hole z = smoothn(y); % smooth data subplot(2,2,1:2), imagesc(y), axis equal off title('Noisy corrupt data') subplot(223), imagesc(z), axis equal off title('Recovered data ...') subplot(224), imagesc(y0), axis equal off title('... compared with the actual data')

Example #4: smooth volumetric data
[x,y,z] = meshgrid(-2:.2:2); xslice = [-0.8,1]; yslice = 2; zslice = [-2,0]; vn = x.*exp(-x.^2-y.^2-z.^2) + randn(size(x))*0.06; subplot(121), slice(x,y,z,vn,xslice,yslice,zslice,'cubic') axis square title('Noisy data') v = smoothn(vn); subplot(122), slice(x,y,z,v,xslice,yslice,zslice,'cubic') axis square title('Smoothed data')

Example #5: smooth a cardioid
t = linspace(0,2*pi,1000);
x = 2*cos(t).*(1-cos(t)) + randn(size(t))*0.1;
y = 2*sin(t).*(1-cos(t)) + randn(size(t))*0.1;
z = smoothn({x,y});
figure
plot(x,y,'r.',z{1},z{2},'k','linewidth',2)
axis equal tight

Example #6: smooth a 3-D parametric curve
t = linspace(0,6*pi,1000);
x = sin(t) + 0.1*randn(size(t));
y = cos(t) + 0.1*randn(size(t));
z = t + 0.1*randn(size(t));
u = smoothn({x,y,z});
figure
plot3(x,y,z,'r.',u{1},u{2},u{3},'k','linewidth',2)
axis tight square

Example #7: smooth a 2-D vector field
[x,y] = meshgrid(linspace(0,1,24)); Vx = cos(2*pi*x+pi/2).*cos(2*pi*y); Vy = sin(2*pi*x+pi/2).*sin(2*pi*y); Vx = Vx + sqrt(0.05)*randn(24,24); % adding Gaussian noise Vy = Vy + sqrt(0.05)*randn(24,24); % adding Gaussian noise I = randperm(numel(Vx)); Vx(I(1:30)) = (rand(30,1)-0.5)*5; % adding outliers Vy(I(1:30)) = (rand(30,1)-0.5)*5; % adding outliers Vx(I(31:60)) = NaN; % missing values Vy(I(31:60)) = NaN; % missing values Vs = smoothn({Vx,Vy},'robust'); % automatic smoothing subplot(121), quiver(x,y,Vx,Vy,2.5), axis square off tight title('Noisy velocity field') subplot(122), quiver(x,y,Vs{1},Vs{2}), axis square off tight title('Smoothed velocity field')

Example #8: smooth a 3-D vector field
Load data and make them noisy
load wind % original 3-D flow % Gaussian noise un = u + randn(size(u))*8; vn = v + randn(size(v))*8; wn = w + randn(size(w))*8; % Add some outliers I = randperm(numel(u)) < numel(u)*0.025; un(I) = (rand(1,nnz(I))-0.5)*200; vn(I) = (rand(1,nnz(I))-0.5)*200; wn(I) = (rand(1,nnz(I))-0.5)*200;
Visualize the noisy flow (see CONEPLOT documentation)
figure, title('Noisy 3-D vectorial flow') xmin = min(x(:)); xmax = max(x(:)); ymin = min(y(:)); ymax = max(y(:)); zmin = min(z(:)); daspect([2,2,1]) xrange = linspace(xmin,xmax,8); yrange = linspace(ymin,ymax,8); zrange = 3:4:15; [cx,cy,cz] = meshgrid(xrange,yrange,zrange); k = 0.1; hcones = coneplot(x,y,z,un*k,vn*k,wn*k,cx,cy,cz,0); set(hcones,'FaceColor','red','EdgeColor','none') hold on wind_speed = sqrt(un.^2 + vn.^2 + wn.^2); hsurfaces = slice(x,y,z,wind_speed,[xmin,xmax],ymax,zmin); set(hsurfaces,'FaceColor','interp','EdgeColor','none') hold off axis tight; view(30,40); axis off camproj perspective; camzoom(1.5) camlight right; lighting phong set(hsurfaces,'AmbientStrength',.6) set(hcones,'DiffuseStrength',.8)

Smooth the noisy flow and display the result
Vs = smoothn({un,vn,wn},'robust');
figure, title('3-D flow smoothed by SMOOTHN')
daspect([2,2,1])
hcones = coneplot(x,y,z,Vs{1}*k,Vs{2}*k,Vs{3}*k,cx,cy,cz,0);
set(hcones,'FaceColor','red','EdgeColor','none')
hold on
wind_speed = sqrt(Vs{1}.^2 + Vs{2}.^2 + Vs{3}.^2);
hsurfaces = slice(x,y,z,wind_speed,[xmin,xmax],ymax,zmin);
set(hsurfaces,'FaceColor','interp','EdgeColor','none')
hold off
axis tight; view(30,40); axis off
camproj perspective; camzoom(1.5)
camlight right; lighting phong
set(hsurfaces,'AmbientStrength',.6)
set(hcones,'DiffuseStrength',.8)

Pick of the week in Matlab FEX
References
Please refer to the two following papers:
- Garcia D. Robust smoothing of gridded data in one and higher dimensions with missing values. Computational Statistics & Data Analysis 2010;54:1167-1178.
- Garcia D. A fast all-in-one method for automated post-processing of PIV data. Exp Fluids 2011;50:1247-1259.
See also
About the author
Damien Garcia, Eng., Ph.D. INSERM researcher Creatis, University of Lyon, France
website: BioméCardio