RMSCAT Remove insignificant scatterers for faster simulations
RMSCAT removes the scatterer that would have little effect on an ultrasound image simulated by SIMUS or SIMUS3.
Contents
Syntax
[X2,Y2,Z2,RC2] = RMSCAT(X,Y,Z,RC,DELAYS,PARAM) removes the scatterers that would have little effect on an ultrasound image simulated by SIMUS or SIMUS3 and of dynamic range 40 dB. The retained scatterers are located at {|X2|,|Y2|,|Z2|} and their reflection coefficients are RC2.
RMSCAT can be used before SIMUS or SIMUS3. After using RMSCAT, you can thus consider running RF = SIMUS(X2,Y2,Z2,RC2,DELAYS,PARAM) instead of RF = SIMUS(X,Y,Z,RC,DELAYS,PARAM). Same for SIMUS3.
[X2,Z2,RC2] = RMSCAT(X,Z,RC,DELAYS,PARAM) assumes that you will use 2-D acoustics in the X-|Z| plane: RF = SIMUS(X2,Z2,RC2,DELAYS,PARAM) or RF = SIMUS(X2,[],Z2,RC2,DELAYS,PARAM).
I = RMSCAT(...) returns the linear indices corresponding to the retained scatterers, i.e. X2 = X(I), Y2 = Y(I),...
[...] = RMSCAT(...,DR) removes the scatterers that would have little effect on a simulated ultrasound of dynamic range DR dB. By default, DR = 40.
Example: Ultrasound image of a carotid
This example shows how you can make simulations faster with RMSCAT
Read a carotid image
I = rgb2gray(imread('carotid.jpg'));
Choose a linear array
param = getparam('L11-5v');
Generate scatterers with GENSCAT
[xs,~,zs,RC] = genscat([5e-2 NaN],param,I,.4);
Simulate an ultrasound image with SIMUS (plane wave)
dels = zeros(1,param.Nelements); % all delays are zero (plane wave) opt.WaitBar = false; tic RF1 = simus(xs,zs,RC,dels,param,opt); t1 = toc; % computation time for SIMUS
Remove some scatterers with RMSCAT
tic
[xs2,zs2,RC2] = rmscat(xs,zs,RC,dels,param);
t2 = toc; % computation time for RMSCAT + SIMUS
Display the scatterers
subplot(211) scatter(xs*1e2,zs*1e2,2,20*log10(RC/max(RC(:))),'filled') ylabel('[cm]') clim([-40 0]) % range = [-40 0] dB axis ij tight equal title([int2str(numel(RC)) ' scatterers']) set(gca,'XColor','none','box','off') subplot(212) scatter(xs2*1e2,zs2*1e2,2,20*log10(RC2/max(RC2(:))),'filled') ylabel('[cm]') clim([-40 0]) % range = [-40 0] dB axis ij tight equal title({'after RMSCAT:',[int2str(numel(RC2)) ' scatterers']}) set(gca,'XColor','none','box','off') colormap(hot)
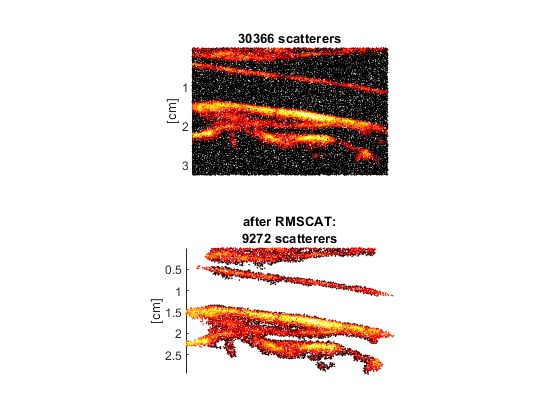
Simulate again for a comparison
tic [RF2,param] = simus(xs2,zs2,RC2,dels,param,opt); t2 = t2 + toc;
Demodulate with RF2IQ
IQ1 = rf2iq(RF1,param.fs,param.fc); IQ2 = rf2iq(RF2,param.fs,param.fc);
Beamform with DAS
[x,z] = meshgrid(linspace(-2e-2,2e-2,256),linspace(eps,3e-2,200)); param.fnumber = []; IQ1b = das(IQ1,x,z,dels,param); IQ2b = das(IQ2,x,z,dels,param);
Create B-mode images with BMODE
I1 = bmode(IQ1b,50); % dynamic range = 50 dB
I2 = bmode(IQ2b,50);
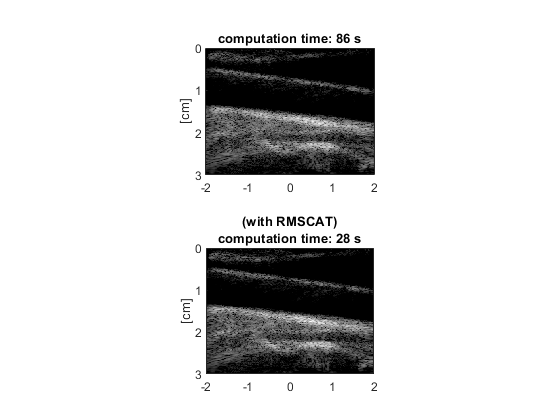
Display the images, and compare
subplot(211) imagesc(x(1,:)*100,z(:,1)*100,I1) ylabel('[cm]') axis equal ij tight title(['computation time: ' int2str(t1) ' s']) subplot(212) imagesc(x(1,:)*100,z(:,1)*100,I2) ylabel('[cm]') axis equal ij tight title({'(with RMSCAT)',['computation time: ' int2str(t2) ' s']}) colormap gray
See also
About the author
Damien Garcia, Eng., Ph.D. INSERM research director Creatis, University of Lyon, France
websites: BioméCardio, MUST
Date modified